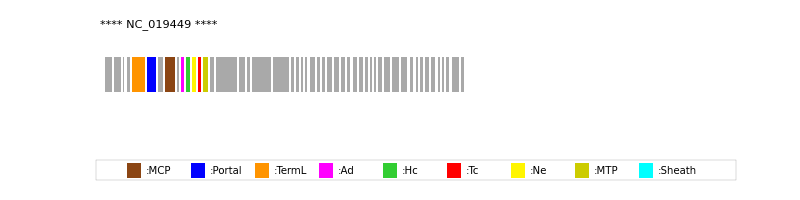
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 8 | lcl|NC_019449.1_cdsid_YP_007002345.1 | HHsearch (proba=100.00%) |
| Portal | 6 | lcl|NC_019449.1_cdsid_YP_007002343.1 | HHsearch (proba=100.00%) |
| TermL | 5 | lcl|NC_019449.1_cdsid_YP_007002342.1 | Blast (e-value=2e-42) |
| MTP | 14 | lcl|NC_019449.1_cdsid_YP_007002351.1 | Blast (e-value=3e-06) |
| Sheath | NOT FOUND | ||
| Ad1 | 10 | lcl|NC_019449.1_cdsid_YP_007002347.1 | HHsearch (proba=99.93%) |
| Hc1 | 11 | lcl|NC_019449.1_cdsid_YP_007002348.1 | HHsearch (proba=100.00%) |
| Ne1 | 12 | lcl|NC_019449.1_cdsid_YP_007002349.1 | HHsearch (proba=97.60%) |
| Tc1 | 13 | lcl|NC_019449.1_cdsid_YP_007002350.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:4456 protein:vir:485 protein:vir:6212 | ST64B P27 phBC6A52 | 20 % 20 % 20 % |
| Portal | protein:vir:105064 protein:vir:5737 protein:vir:102080 | phiKO2 PY54 Fah | 23 % 22 % 21 % |
| TermL | protein:vir:81178 protein:vir:78630 protein:vir:94427 | Geobacillus virus E2 tp310-2 47 | 26 % 25 % 25 % |
| MTP | protein:vir:102037 protein:vir:102845 protein:vir:107619 | Fah Cherry Gamma | 23 % 23 % 23 % |
| Ad1 | protein:vir:1271 protein:vir:81159 protein:vir:102083 | phi-105 Geobacillus virus E2 Fah | 26 % 23 % 22 % |
| Hc1 | protein:vir:100134 protein:vir:81177 protein:vir:105035 | phi1026b Geobacillus virus E2 phiKO2 | 25 % 25 % 24 % |
| Ne1 | protein:vir:100243 protein:vir:80362 protein:vir:1273 | Bcep176 phi644-2 phi-105 | 22 % 23 % 23 % |
| Tc1 | protein:vir:1387 protein:vir:102086 protein:vir:102888 | phi3626 Fah Cherry | 28 % 22 % 22 % |