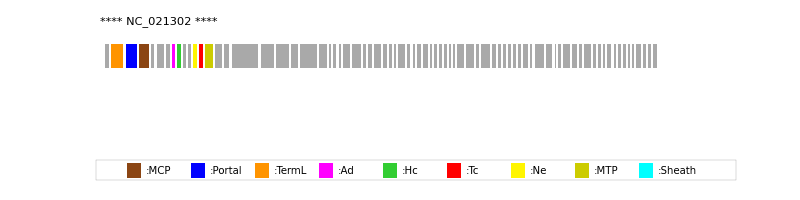
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 4 | lcl|NC_021302.1_cdsid_YP_008051229.1 | HHsearch (proba=100.00%) |
| Portal | 3 | lcl|NC_021302.1_cdsid_YP_008051228.1 | HHsearch (proba=100.00%) |
| TermL | 2 | lcl|NC_021302.1_cdsid_YP_008051227.1 | Blast (e-value=1e-101) |
| MTP | 14 | lcl|NC_021302.1_cdsid_YP_008051239.1 | Blast (e-value=9e-37) |
| Sheath | NOT FOUND | ||
| Ad1 | 8 | lcl|NC_021302.1_cdsid_YP_008051233.1 | HHsearch (proba=100.00%) |
| Hc1 | 9 | lcl|NC_021302.1_cdsid_YP_008051234.1 | HHsearch (proba=100.00%) |
| Ne1 | 12 | lcl|NC_021302.1_cdsid_YP_008051237.1 | HHsearch (proba=100.00%) |
| Tc1 | 13 | lcl|NC_021302.1_cdsid_YP_008051238.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Proteins Superfamilies | Observed Intergene Distance | Expected Intergene Distance |
|---|---|---|
| Tc1<->Hc1 | 4 | 3 |
| Ne1<->Hc1 | 3 | 2 |
| Tc1<->Ad1 | 5 | 4 |
| Ne1<->Ad1 | 4 | 3 |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:79548 | cdtI | 32 % |
| Portal | protein:vir:108215 protein:vir:78161 protein:vir:95254 | Giles Min1 Felix 01 | 43 % 40 % 22 % |
| TermL | protein:vir:108214 protein:vir:80678 protein:vir:99071 | Giles PA6 Wildcat | 41 % 24 % 23 % |
| MTP | protein:vir:101656 protein:vir:7861 protein:vir:8436 | 244 CJW1 Omega | 30 % 30 % 29 % |
| Ad1 | protein:vir:108221 protein:vir:9761 protein:vir:9576 | Giles 315.3 SM1 | 28 % 23 % 23 % |
| Hc1 | protein:vir:108230 protein:vir:8328 protein:vir:8105 | Giles Corndog Che9c | 37 % 27 % 18 % |
| Ne1 | protein:vir:8432 protein:vir:8106 protein:vir:8330 | Omega Che9c Corndog | 34 % 30 % 29 % |
| Tc1 | protein:vir:8331 protein:vir:102609 protein:vir:105826 | Corndog Llij PMC | 33 % 30 % 30 % |