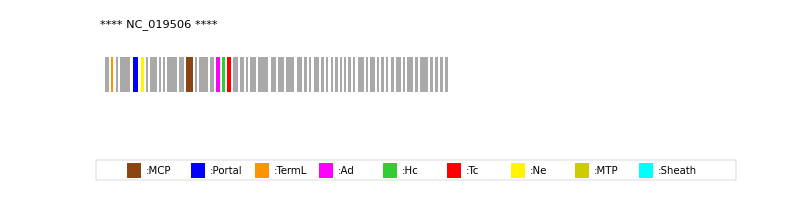
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 13 | lcl|NC_019506.1_cdsid_YP_007005028.1 | HHsearch (proba=100.00%) |
| Portal | 5 | lcl|NC_019506.1_cdsid_YP_007005020.1 | HHsearch (proba=99.94%) |
| TermL | 2 | lcl|NC_019506.1_cdsid_YP_007005017.1 | Blast (e-value=2e-14) |
| MTP | NOT FOUND | ||
| Sheath | NOT FOUND | ||
| Ad1 | 17 | lcl|NC_019506.1_cdsid_YP_007005032.1 | HHsearch (proba=97.32%) |
| Hc1 | 18 | lcl|NC_019506.1_cdsid_YP_007005033.1 | HHsearch (proba=89.66%) |
| Ne1 | 6 | lcl|NC_019506.1_cdsid_YP_007005021.1 | HHsearch (proba=99.85%) |
| Tc1 | 19 | lcl|NC_019506.1_cdsid_YP_007005034.1 | HHsearch (proba=76.35%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Proteins Superfamilies | Observed Intergene Distance | Expected Intergene Distance |
|---|---|---|
| Ne1<->Hc1 | 12 | 2 |
| Ne1<->Ad1 | 11 | 3 |
| Ne1<->Tc1 | 13 | 2 |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:102605 protein:vir:105822 protein:vir:7990 | Llij PMC Che8 | 31 % 31 % 30 % |
| Portal | protein:vir:94666 | mu1/6 | 22 % |
| TermL | protein:vir:97448 protein:vir:95058 protein:vir:94499 | 92 X2 88 | 45 % 45 % 45 % |
| Ad1 | protein:vir:78068 protein:vir:105327 protein:vir:107119 | P35 PH15 CNPH82 | 19 % 26 % 26 % |
| Hc1 | protein:vir:5977 protein:vir:100134 protein:vir:80342 | SPP1 phi1026b phi644-2 | 13 % 11 % 14 % |
| Ne1 | protein:vir:102085 protein:vir:102875 protein:vir:107568 | Fah Cherry Gamma | 29 % 29 % 29 % |
| Tc1 | protein:vir:10368 protein:vir:1244 | Xp10 phi ETA | 15 % 13 % |