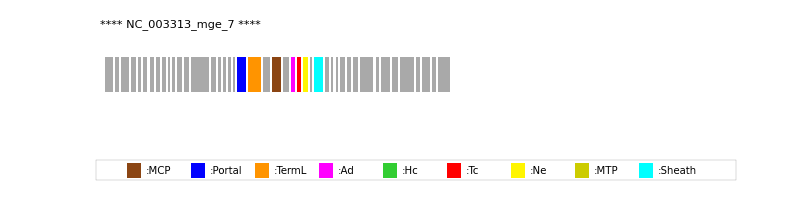
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 23 | lcl|Aclame:protein:vir:270|NCBI_annot:putative | HHsearch (proba=100.00%) |
| Portal | 20 | lcl|Aclame:protein:vir:267|NCBI_annot:putative | HHsearch (proba=100.00%) |
| TermL | 21 | lcl|Aclame:protein:vir:268|NCBI_annot:putative | Blast (e-value=0) |
| MTP | NOT FOUND | ||
| Sheath | 29 | lcl|Aclame:protein:vir:276|NCBI_annot:putative | HHsearch (proba=100.00%) |
| Ad1 | 25 | lcl|Aclame:protein:vir:272|NCBI_annot:putative | HHsearch (proba=100.00%) |
| Hc1 | NOT FOUND | ||
| Ne1 | 27 | lcl|Aclame:protein:vir:274|NCBI_annot:putative | HHsearch (proba=100.00%) |
| Tc1 | 26 | lcl|Aclame:protein:vir:273|NCBI_annot:hypothetical | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:270 protein:vir:78777 protein:vir:1153 | K139 phiO18P phi CTX | 100 % 48 % 32 % |
| Portal | protein:vir:267 protein:vir:98853 protein:vir:78749 | K139 F108 phiO18P | 100 % 49 % 47 % |
| TermL | protein:vir:268 protein:vir:78781 protein:vir:3744 | K139 phiO18P HP1 | 100 % 57 % 50 % |
| Sheath | protein:vir:276 protein:vir:78782 protein:vir:3788 | K139 phiO18P HP2 | 100 % 43 % 41 % |
| Ad1 | protein:vir:272 protein:vir:78780 protein:vir:3748 | K139 phiO18P HP1 | 100 % 44 % 37 % |
| Ne1 | protein:vir:274 protein:vir:78755 protein:vir:98860 | K139 phiO18P F108 | 100 % 26 % 24 % |
| Tc1 | protein:vir:273 protein:vir:78779 protein:vir:3786 | K139 phiO18P HP2 | 100 % 38 % 24 % |