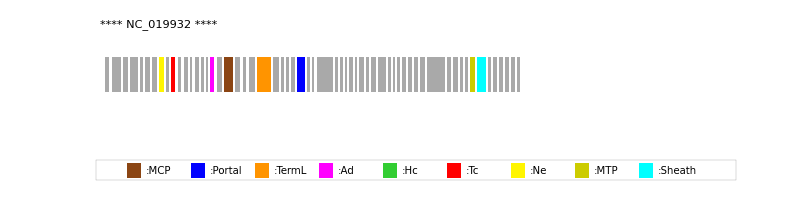
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 19 | lcl|NC_019932.1_cdsid_YP_007238029.1 | HHsearch (proba=100.00%) |
| Portal | 28 | lcl|NC_019932.1_cdsid_YP_007238038.1 | HHsearch (proba=100.00%) |
| TermL | 23 | lcl|NC_019932.1_cdsid_YP_007238033.1 | Blast (e-value=0) |
| MTP | 53 | lcl|NC_019932.1_cdsid_YP_007238063.1 | Blast (e-value=2e-61) |
| Sheath | 54 | lcl|NC_019932.1_cdsid_YP_007238064.1 | HHsearch (proba=100.00%) |
| Ad1 | 17 | lcl|NC_019932.1_cdsid_YP_007238027.1 | HHsearch (proba=100.00%) |
| Hc1 | NOT FOUND | ||
| Ne1 | 8 | lcl|NC_019932.1_cdsid_YP_007238018.1 | HHsearch (proba=100.00%) |
| Tc1 | 10 | lcl|NC_019932.1_cdsid_YP_007238020.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Proteins Superfamilies | Observed Intergene Distance | Expected Intergene Distance |
|---|---|---|
| Tc1<->Ad1 | 7 | 4 |
| Ne1<->Ad1 | 9 | 4 |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:5694 protein:vir:2016 protein:vir:6061 | L-413C P2 WPhi | 74 % 74 % 73 % |
| Portal | protein:vir:2013 protein:vir:6058 protein:vir:5691 | P2 WPhi L-413C | 69 % 69 % 69 % |
| TermL | protein:vir:98555 protein:vir:2014 protein:vir:6059 | PSP3 P2 WPhi | 72 % 71 % 71 % |
| MTP | protein:vir:2036 protein:vir:6080 protein:vir:5712 | P2 WPhi L-413C | 66 % 66 % 65 % |
| Sheath | protein:vir:1845 protein:vir:98553 protein:vir:6079 | 186 PSP3 WPhi | 80 % 79 % 78 % |
| Ad1 | protein:vir:1831 protein:vir:98558 protein:vir:2018 | 186 PSP3 P2 | 51 % 49 % 49 % |
| Ne1 | protein:vir:1838 protein:vir:98557 protein:vir:2026 | 186 PSP3 P2 | 65 % 61 % 45 % |
| Tc1 | protein:vir:2025 protein:vir:5702 protein:vir:98561 | P2 L-413C PSP3 | 59 % 58 % 57 % |