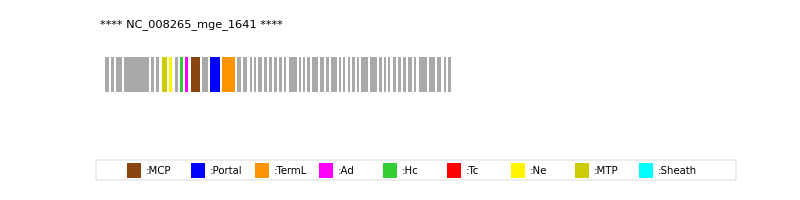
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 12 | lcl|Aclame:protein:vir:102119|NCBI_annot:phage | HHsearch (proba=100.00%) |
| Portal | 14 | lcl|Aclame:protein:vir:102118|NCBI_annot:phage | HHsearch (proba=100.00%) |
| TermL | 15 | lcl|Aclame:protein:vir:102145|NCBI_annot:phage | Blast (e-value=0) |
| MTP | 7 | lcl|Aclame:protein:vir:102112|NCBI_annot:phage | Blast (e-value=1e-123) |
| Sheath | NOT FOUND | ||
| Ad1 | 11 | lcl|Aclame:protein:vir:102158|NCBI_annot:uncharacterized | HHsearch (proba=100.00%) |
| Hc1 | 10 | lcl|Aclame:protein:vir:102144|NCBI_annot:phage | HHsearch (proba=100.00%) |
| Ne1 | 8 | lcl|Aclame:protein:vir:102154|NCBI_annot:phage | HHsearch (proba=100.00%) |
| Tc1 | NOT FOUND | ||
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:102119 protein:vir:81160 protein:vir:102082 | phiSM101 Geobacillus virus E2 Fah | 100 % 35 % 33 % |
| Portal | protein:vir:102118 protein:vir:102080 protein:vir:102855 | phiSM101 Fah Cherry | 100 % 56 % 56 % |
| TermL | protein:vir:102145 protein:vir:81178 protein:vir:1379 | phiSM101 Geobacillus virus E2 phi3626 | 100 % 36 % 35 % |
| MTP | protein:vir:102112 protein:vir:102037 protein:vir:102845 | phiSM101 Fah Cherry | 100 % 23 % 23 % |
| Ad1 | protein:vir:102158 protein:vir:81159 protein:vir:102083 | phiSM101 Geobacillus virus E2 Fah | 100 % 48 % 44 % |
| Hc1 | protein:vir:102144 protein:vir:1385 protein:vir:1890 | phiSM101 phi3626 HK022 | 100 % 29 % 28 % |
| Ne1 | protein:vir:102154 protein:vir:80362 protein:vir:102085 | phiSM101 phi644-2 Fah | 100 % 27 % 27 % |