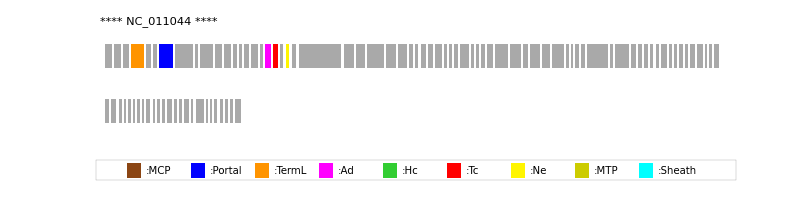
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | NOT FOUND | ||
| Portal | 7 | lcl|NC_011044.1_cdsid_YP_002003846.1 | HHsearch (proba=100.00%) |
| TermL | 4 | lcl|NC_011044.1_cdsid_YP_002003843.1 | Blast (e-value=0) |
| MTP | NOT FOUND | ||
| Sheath | NOT FOUND | ||
| Ad1 | 18 | lcl|NC_011044.1_cdsid_YP_002003857.1 | HHsearch (proba=100.00%) |
| Hc1 | NOT FOUND | ||
| Ne1 | 21 | lcl|NC_011044.1_cdsid_YP_002003860.1 | HHsearch (proba=100.00%) |
| Tc1 | 19 | lcl|NC_011044.1_cdsid_YP_002003858.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| Portal | protein:vir:106027 protein:vir:102426 protein:vir:99088 | Cooper Pipefish Qyrzula | 90 % 56 % 55 % |
| TermL | protein:vir:106049 protein:vir:97894 protein:vir:107494 | Cooper Orion PG1 | 96 % 67 % 67 % |
| Ad1 | protein:vir:106038 protein:vir:97979 protein:vir:107532 | Cooper Orion PG1 | 79 % 61 % 61 % |
| Ne1 | protein:vir:106041 protein:vir:97982 protein:vir:107545 | Cooper Orion PG1 | 86 % 71 % 71 % |
| Tc1 | protein:vir:106039 protein:vir:97980 protein:vir:107514 | Cooper Orion PG1 | 90 % 63 % 63 % |