 InterEvolAlign
InterEvolAlign
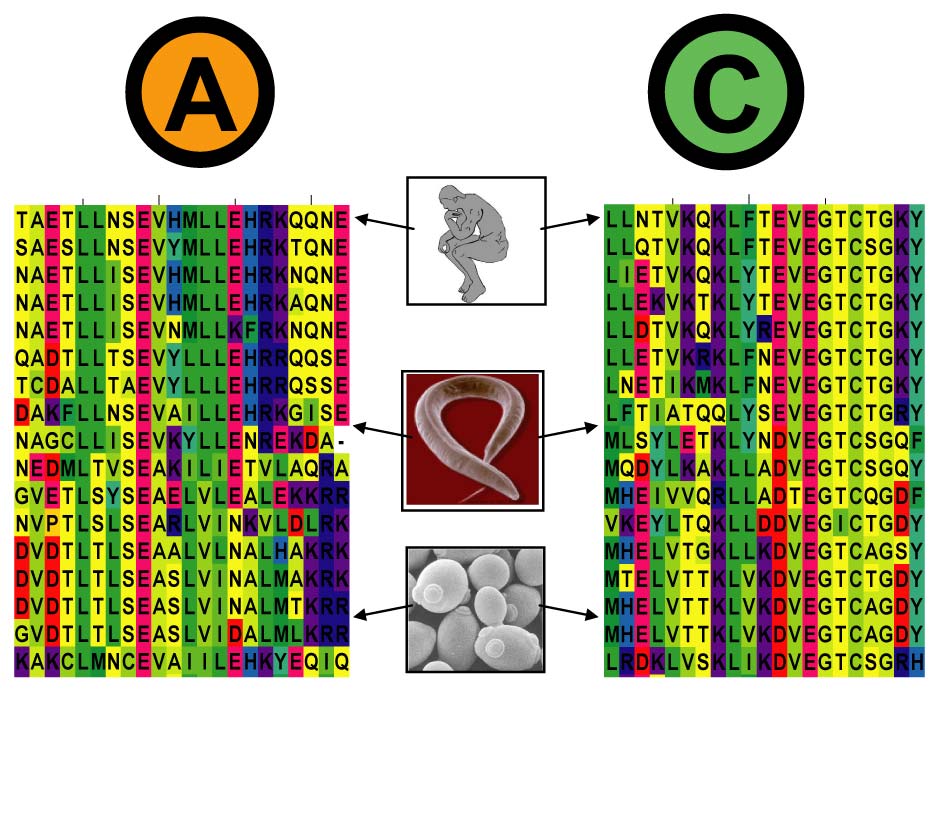
Build alignments and query InterEvol database
From 1 or 2 sequences of interacting partners:
- build 2 multiple sequence alignments with the same species ordered in each
- query the InterEvol database with alignments using profile-profile comparison method

