This website is free and open to all users and there is no
login requirement.
- PyMOL is a popular
program for protein structure visualisation.
- We developed the InterEvol PyMOL plugin to perform
structure-oriented exploration of the sequence alignments.
- Use the 3D structure of a complex (from a PDB file or a
model)
- Read the alignments of every chain made by the InterEvolAlign
server or downloaded from InterEvol
database
- Interactively analyse the selected residues in Pymol
and their columns extracted from the sequence alignment.
- First, you need to download
the file InterEvolPyMOLPlugin v1.0. For any
bug or question please contact us.
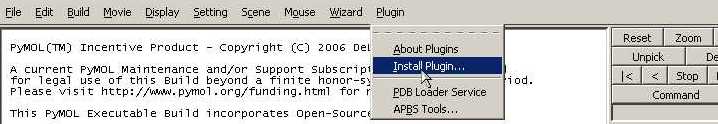
1 - Install the InterEvol PyMOL Plugin
- Open your Pymol program
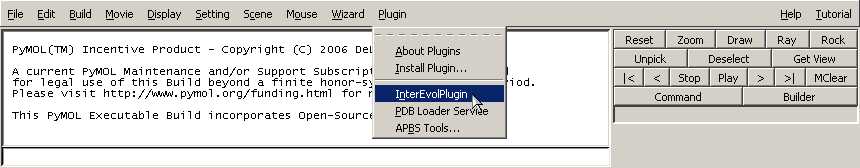
- Click on Plugin>Install Plugin... on the top bar. (Admin
rights might be required depending the location of your PyMOL
directory).
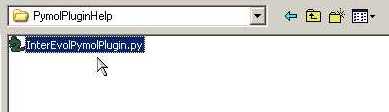
- Select InterEvolPymolPlugin.py
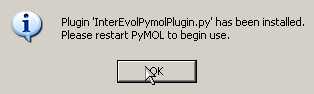
- Once the plugin installed, you need to quit PyMOL
(after clicking
OK).
2 - How to use the InterEvol Pymol Plugin
Loading the PDB and the
alignment files
- Open a pymol session and directly activates the plugin.
- Click on Plugin>InterEvolPlugin.
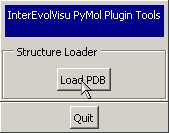
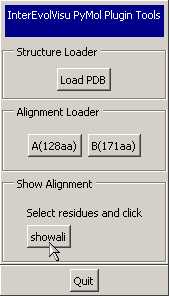
- The following "InterEvol control panel" pops
up. Click "Load PDB" to
open your pdb file
- For the example we will use the structure of the human
Rpb4-Rpb7 complex (PDB code : 2c35.pdb)
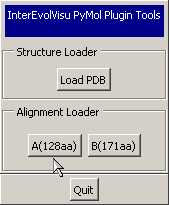
- The panel of the "InterEvol plugin panel" now
displays all the chains found in the PDB (with more than 30 residues)
as buttons than can be used to load the alignments. In the main pymol
window the structure is opened
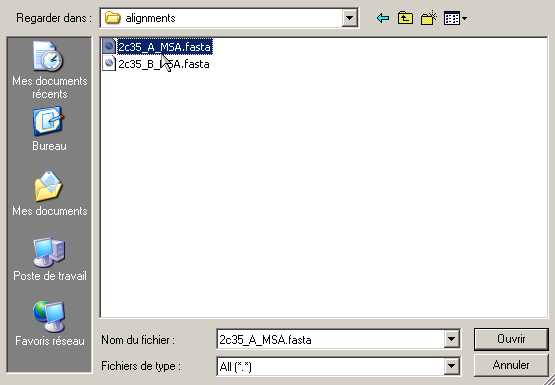
- Load your alignments of interest by selecting the
corresponding file with the "Alignment Loader"
buttons. Here, we successively open 2c35_A_MSA.fasta
and 2c35_B_MSA.fasta for chain A and B,
respectively.
- A new button appears in the "InterEvol
plugin panel".
You can now select a set of residues of interest with the standard
selection tools of Pymol and watch their alignment by clicking the "showali"
button.
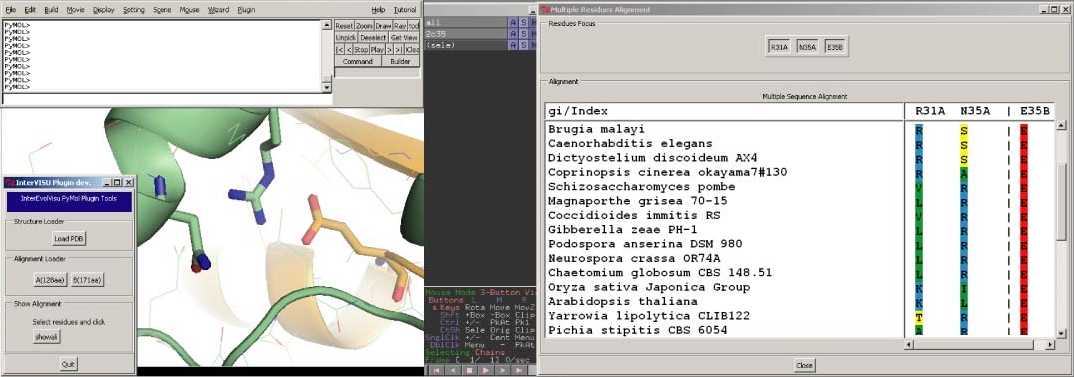
Analyzing an example :
the
R31A-E35B salt-bridge in Rpb4-Rpb7 complex (2c35)
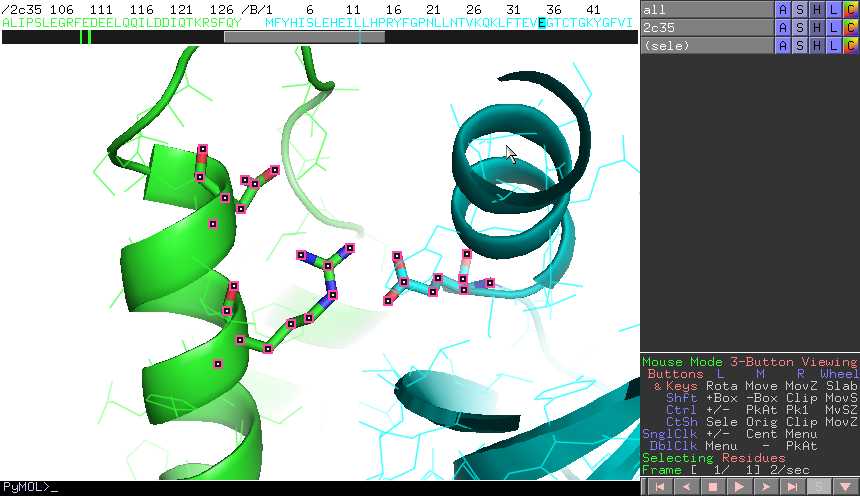
- For instance, let's focus on an interesting site at the
interface of the complex. In chain B, E35
is involved in a buried salt bridge with R31 in chain A.
We will select both residues plus another residue, N35 in
chain A, a structural neighbour of R31.
- Once selected, click on the "showali"
button in the InterEvol plugin panel. You can also run this tool by
typing in the main PyMOL window : "PyMOL> showali (sele)"
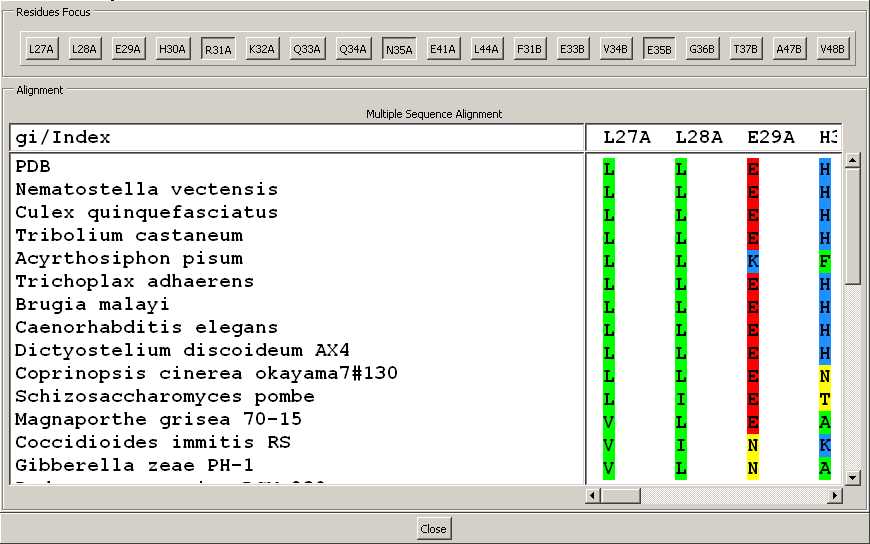
- The following window pops up. You can expand the window
to see all the columns or use the scroll on the right and bottom.
- On top, the buttons "Residues Focus",
helps you to focus back in the structure viewer panel on a restricted
set of residues (see
below).
- At the bottom, the alignment is provided only for the
selected residues. are labelled, if the alignment was made
using InterEvolAlign
server or follow its format, otherwise the gis or indexes are
provided.

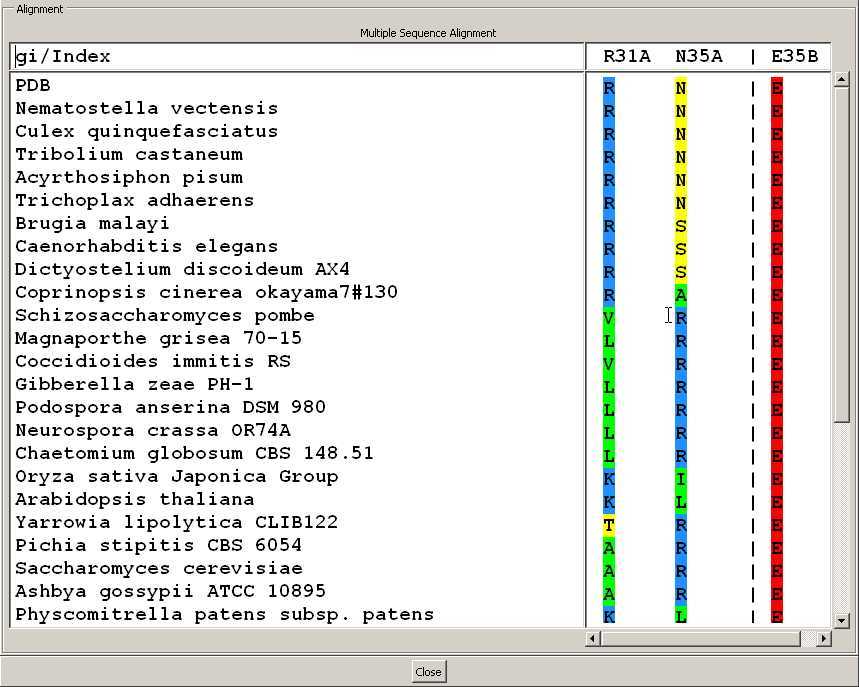
- We can rapidly see that the salt-bridge
R31_chainA:E35_chainB was not conserved throughout eukaryotes
evolution.
- Although E35 was kept invariant,
- R31 was substituted either by a hydrophobic or a polar
residue.
- How could the buried E35 be then accomodated within
the core of the interface ?
- The features of the N35_chainA position
suggest that it may play a compensatory role. It switched to a
basic residue everytime the basic property at position 31 was lost. In
particular, in S. cerevisiae, N35 is substitued into an arginine.
- Thanks to the InterEvol
database browser we can check whether a structural interolog
of 2c35 is available :
- A set of four non redundant structural interologs is
available from eukaryotes but also from archaea.
- 1y14, is the structure of the S.
cervisiae Rpb4-Rpb7 complex sharing about 40%
identity with their human orthologs.
- Interestingly, the 1y14.pdb
structure highlights how the compensation for the loss of salt-bridge
with E35 did occur.
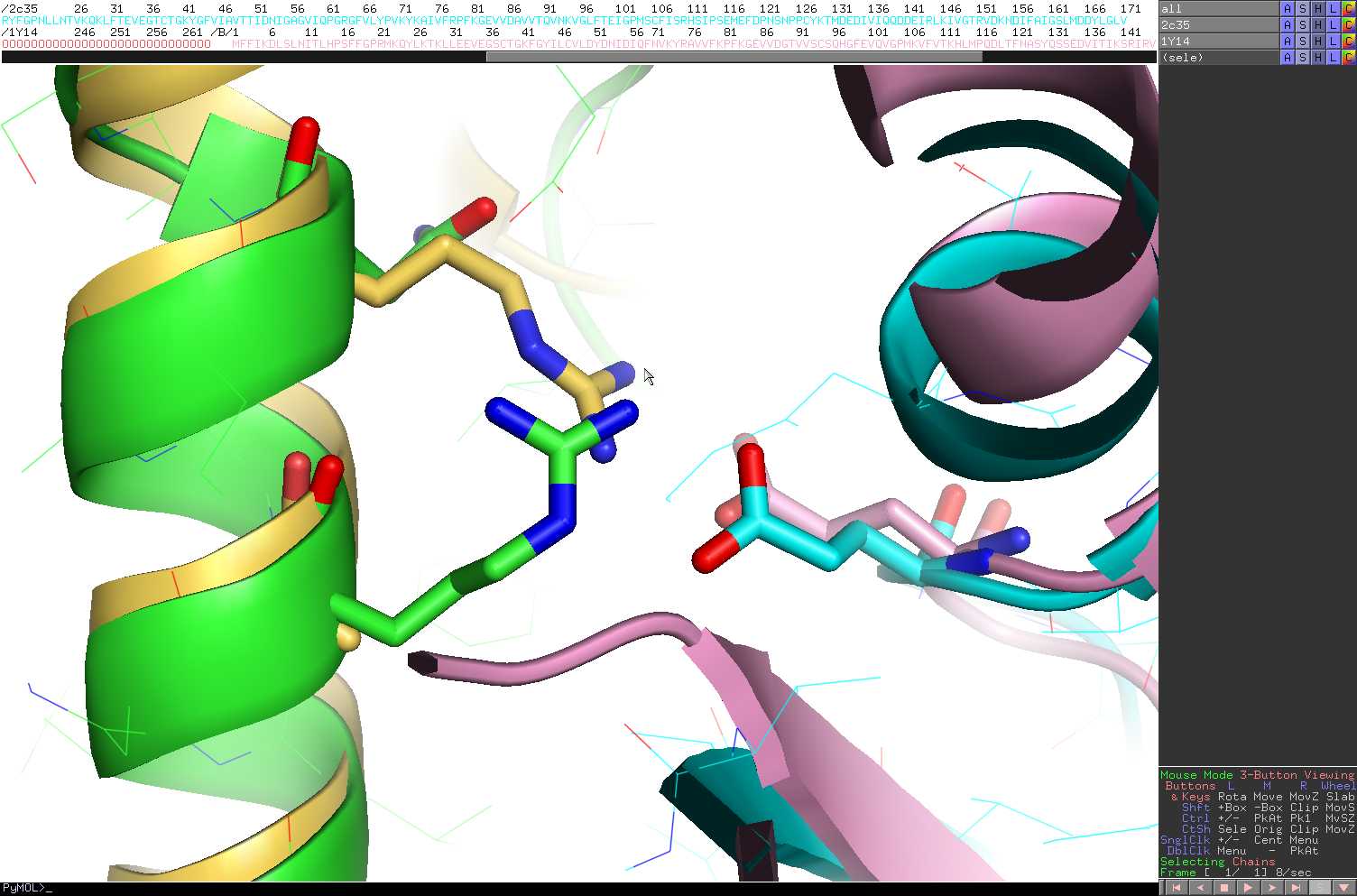
- In this figure the human Rpb4-Rpb7 complex is
shown in green and cyan while the S. cerevisiae is shown in yellow and
pink.
- It confirms the interplay between positions
31_chainA and 35_chainA to neutralize for the buried charge of
E35_chainB.
Cross-talking
between alignments and 3D structure
- In the previous example, only three residues were
selected at first. Many more can be initially selected and the "Residues
Focus" buttons can then help targeting those residues likely
to play compensatory roles in the neighbourhood of E35.
- Buttons can be either activated or disactivated
which toggle in the structure viewer specific zoom and stick
representation
on the residues activated in the "Residues Focus"
panel. All the other residues are automatically shown as simple lines,
allowing a rapid and interactive analysis of the coevolution at
different sites and among different species.
- New selections can then be made and the whole
process using the "showali" button be repeated. Every
time the latter is clicked, a new alignment window will be opened.
3 - Uninstall the Plugin
- To Uninstall InterEvol PyMOL plugin, close the
PyMol program.
- Delete the files <PYMOL_DIR>/modules/pmg_tk/startup/InterEvolPymolPlugin.py
and <PYMOL_DIR>/modules/pmg_tk/startup/InterEvolPymolPlugin.pyc
within the main PyMOL directory.
- If your PyMOL was installed by root/admin, you will
have to
uninstall the Plugin with root/admin priviledge.
- Done !