| InterEvol help |
Three tools available with the InterEvol database
1 - General Scope and Methods.
2 - Browsing of the InterEvol Database.
3 - Building Interolog Multiple Sequence Alignments.
4 - Coupled Analysis of Sequence Alignments and 3D structures at Protein Complex Interfaces.
1 - General Scope and Methods
- The InterEvol server is designed to combine structural and evolutionary analyses of protein complexes interfaces.
- It allows three levels of analysis either through the
structural dimension (InterEvolDB), the sequences alignments one
(InterEvolAlign) and through the combined analysis of both information
through a dedicated PyMOL plugin (InterEvolPymol plugin).
- 36028 homomeric and heteromeric assemblies available in the PDB and involving different proteins were automatically analyzed and are regularly updated.
- 3996 heteromeric non redundant interfaces
between two chains were identified (redundancy threshold at 70 %
identity, one complex in the PDB can contain several interfaces)
- PDB chains and the interface of their complexes were systematically compared and clustered so as to capture :
- Structural homologs, which share similar structures
and common evolutionary properties.
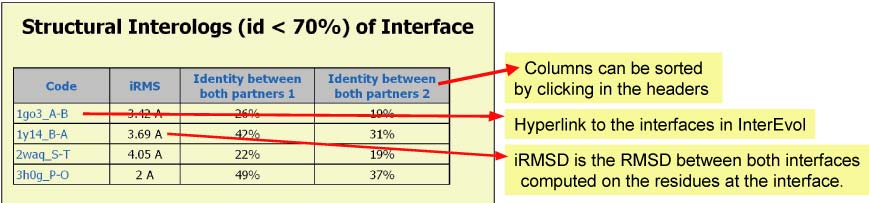
- Structural interologs, i.e. structures of complexes between partners whose homologs also had their structure in complex resolved.
- The characteristics of these interface were analyzed (redundancy filters, sequence and structural divergence, topology, biological/non biological, obligate/non obligate features, etc...) and are available through the InterEvol database browser.
- 1176 homomeric and 520 heteromeric distinct groups of structural interologs provide a wealth of information about the coevolution events at complexes interfaces (one group can contains at least two non redundant interfaces, but can gather more than seven members).
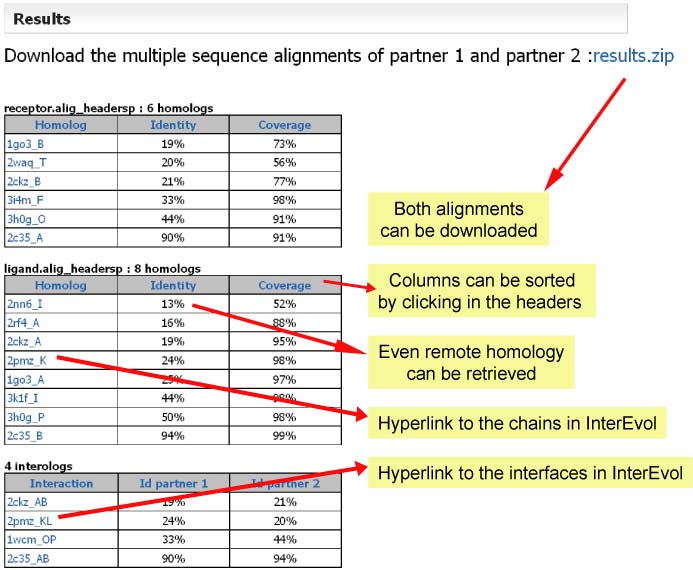
- Additionally, the multiple sequence alignments of the various chains in the InterEvol database were computed so as to provide for every interface an alignment for each partner 1 and 2 with the same species mirroring each other in the alignment.
- The InterEvolAlign server offers the users to generate tailored alignments containing common species for both partners. It was used to generate those of the InterEvol database.
- Visualization can be performed with a dedicated PyMOL plugin.
2 - Browsing of the InterEvol Database
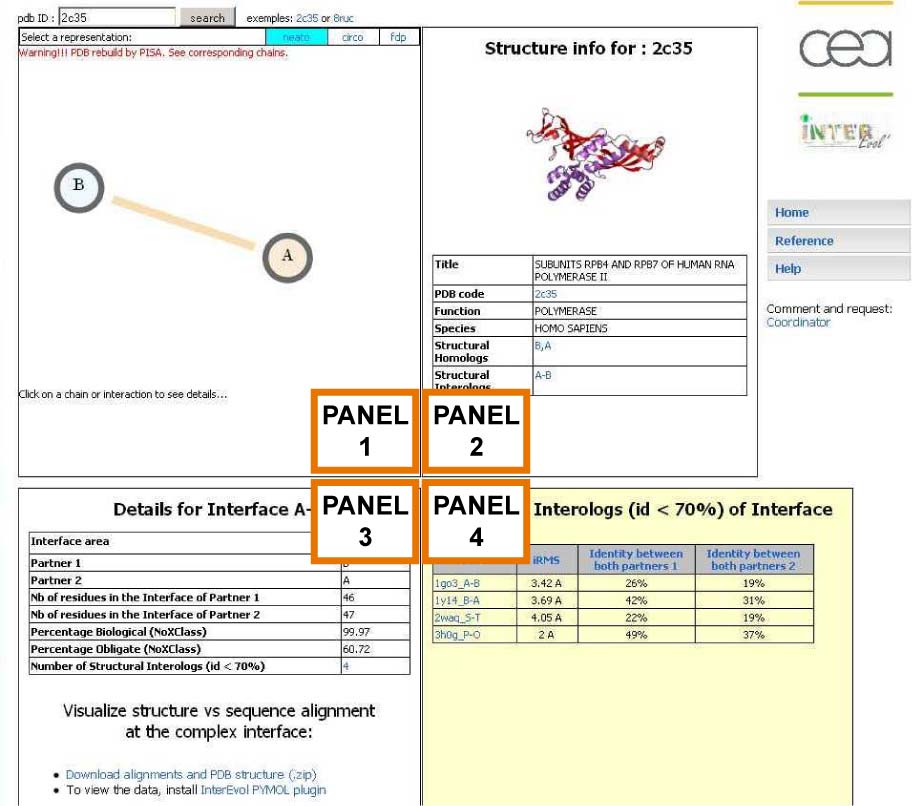
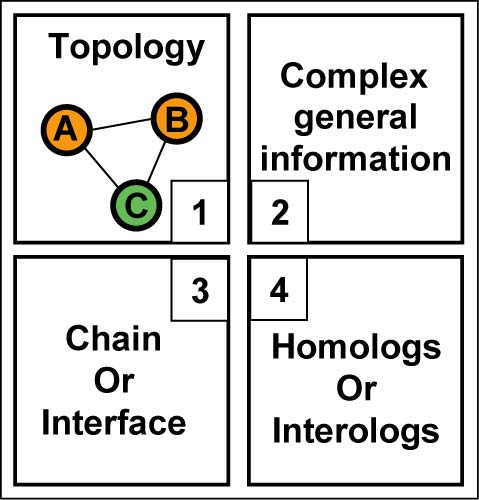
- For every hetero-complex of the PDB, the InterEvol database provides key informations organized in a 4 panel format.
- Panel 1 : Reports the topology of the contacting chains. Nodes (chains) and edges (interfaces) can be clicked to focus the analysis on every part of the PDB.
- Panel 2 : Provide general information about the function of the complex and of its species.
- Panel 3 : Summarizes the information about the chain or the interface selected in Panel 1.
- Panel 4 :
Lists all the homologous chains and structural interologs and
interconnects them through web links.
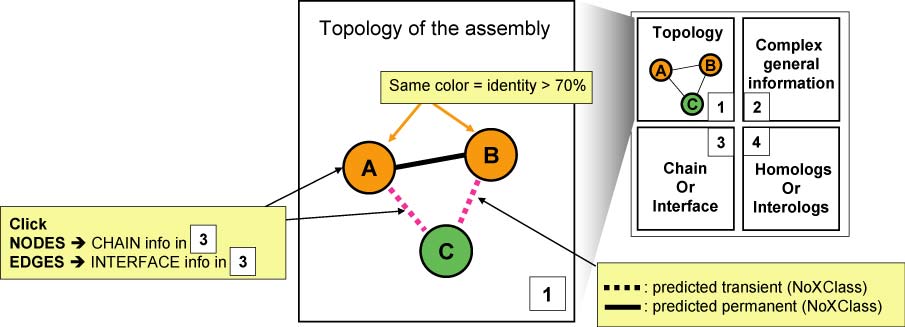
- Three different topological representations can be selected. Chains (nodes) and heteromeric interfaces (edges) can be clicked to explore the complex components.
- The assembly presented is either the assembly as assigned by authors in the original PDB file or derived from PISA quaternary structure generation method.
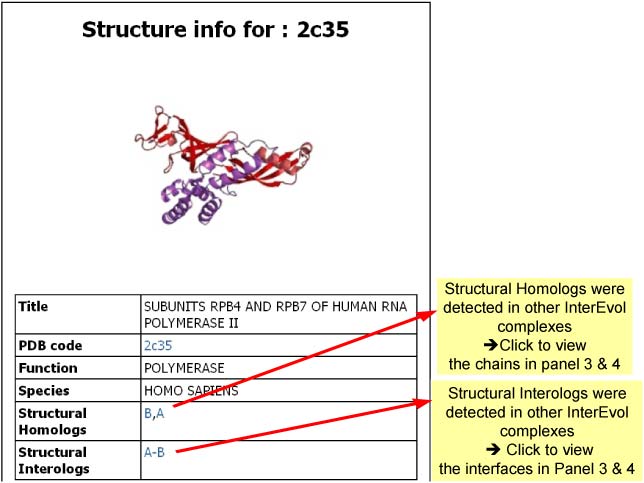
Example for a binary complex (2c35)
Overview of the InterEvol information sheet for a given PDB
Detailed description of every panel is provided below Panel 2 : General Structural Information
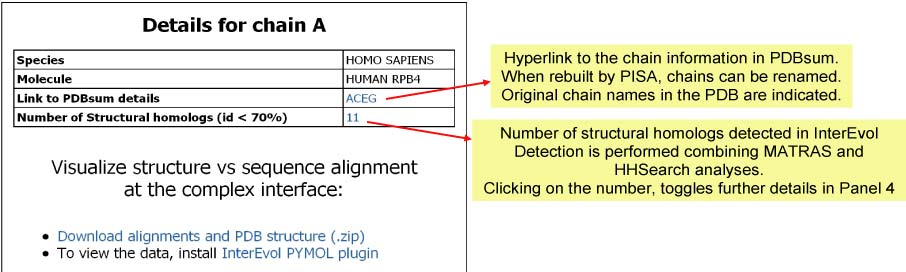
Panel 3a : If a NODE (CHAIN)
is clicked in Panel 1
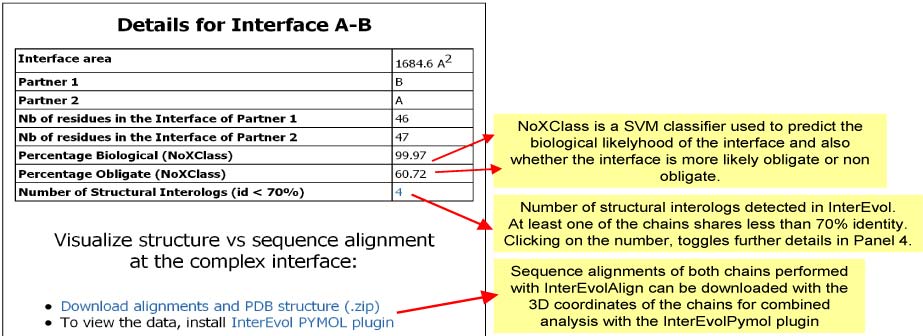
Panel 3b : If an EDGE (INTERFACE) was clicked in Panel 1
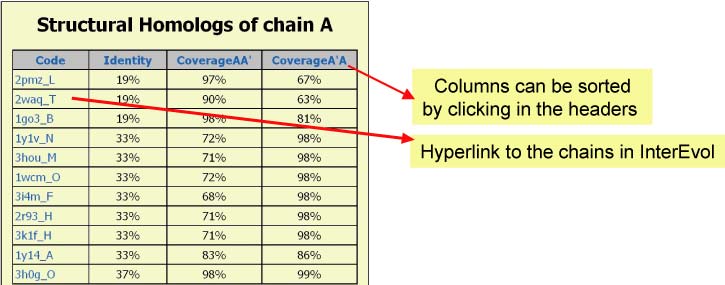
Panel 4a : If a NODE (CHAIN) was clicked in Panel 1
Panel 4b : If an EDGE (INTERFACE) was clicked in Panel 1
3 - Building Interolog Mulltiple Sequence Alignments.
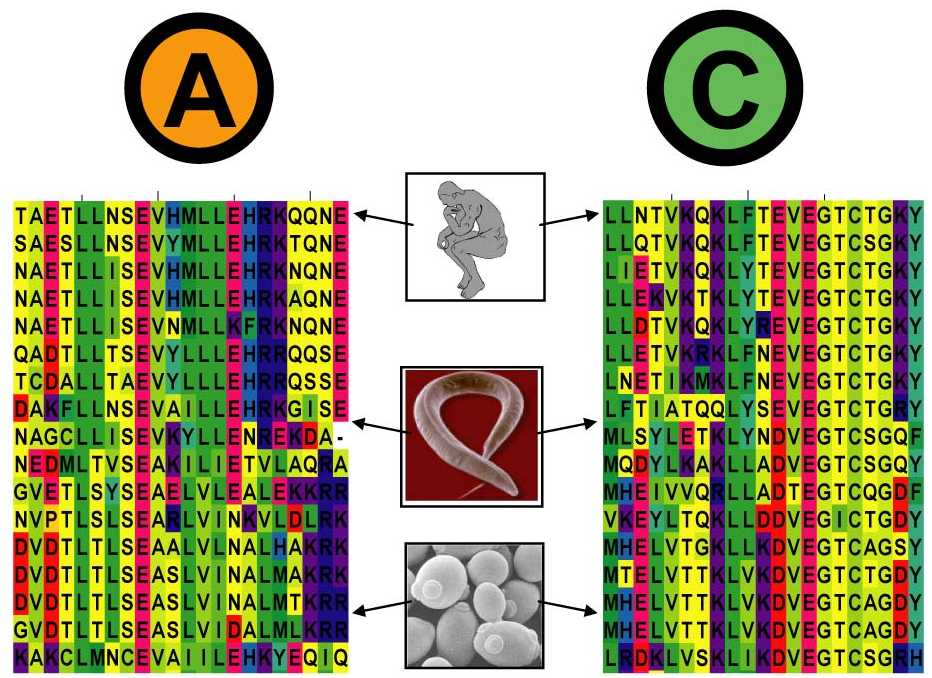
InterEvolAlign server, is designed to generate
two multiple sequence alignments
for the sequences of two partners
known to interact throughout evolution
two multiple sequence alignments
for the sequences of two partners
known to interact throughout evolution
- The search comprises two steps :
1. Generation of a multiple sequence alignment for both sequences with mirroring common species.
2. Search for homologs and remote homologs for every alignment against the InterEvol database using HHsearch.
2. Search for homologs and remote homologs for every alignment against the InterEvol database using HHsearch.
- The InterEvolAlign tool can be useful in at least three configurations :
- You are interested in the alignment of interologs (homologs that kept interacting over evolution) for a given pair of sequences
- You work with a structural model of a complex and you want to get an alignment for both partners to analyze the interface with the InterEvol Pymol Plugin
- You work with a structure that is not available in the InterEvol database.
- The output page is sent via email.
- Typical search will last about 5 minutes depending on the server load.
1. Multiple sequence alignments
- Both generated alignments
are restricted to homologous sequences found in the same species. The
species appear in the same order in both alignments.
- Redundancy can be filtered (default: 90 % seq identity) and the search protocol can be tuned.
- The divergence of the retrieved sequences can be controled so that every sequence share at least one homologous with more than a specific threshold of percentage identity.
- Only one sequence for every species is selected.
2. Searching against the InterEvol database
- Once the multiple sequence alignments are generated, they are used to search for homologous chains in InterEvol using the profile-profile comparison HHSearch.
- The server also searches for Structural Interologs that have been already analyzed in the InterEvol database.
Example of an output obtained with the examples provided in the query page