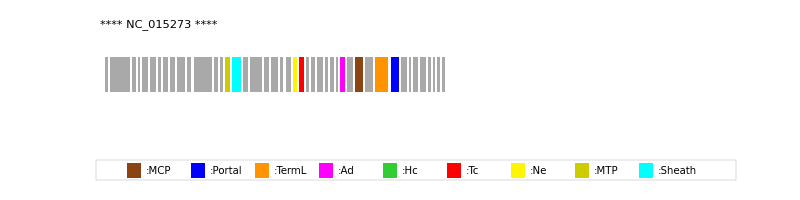
| Protein Superfamily | Index in Query Genome | Sequence Header | Detection Method |
|---|---|---|---|
| MCP | 33 | lcl|NC_015273.1_cdsid_YP_004306877.1 | HHsearch (proba=100.00%) |
| Portal | 36 | lcl|NC_015273.1_cdsid_YP_004306880.1 | HHsearch (proba=100.00%) |
| TermL | 35 | lcl|NC_015273.1_cdsid_YP_004306879.1 | Blast (e-value=0) |
| MTP | 15 | lcl|NC_015273.1_cdsid_YP_004306859.1 | Blast (e-value=5e-63) |
| Sheath | 16 | lcl|NC_015273.1_cdsid_YP_004306860.1 | HHsearch (proba=100.00%) |
| Ad1 | 31 | lcl|NC_015273.1_cdsid_YP_004306875.1 | HHsearch (proba=100.00%) |
| Hc1 | NOT FOUND | ||
| Ne1 | 23 | lcl|NC_015273.1_cdsid_YP_004306867.1 | HHsearch (proba=100.00%) |
| Tc1 | 24 | lcl|NC_015273.1_cdsid_YP_004306868.1 | HHsearch (proba=100.00%) |
| Ad2 | NOT FOUND | ||
| Hc2 | NOT FOUND | ||
| Tc2 | NOT FOUND | ||
| Ad3 | NOT FOUND | ||
| Hc3 | NOT FOUND | ||
| Ad4 | NOT FOUND |
| Proteins Superfamilies | Observed Intergene Distance | Expected Intergene Distance |
|---|---|---|
| Tc1<->Ad1 | 7 | 4 |
| Ne1<->Ad1 | 8 | 4 |
| Protein Superfamily | Corresponding Protein in Aclame | Aclame Phage | Seq. Identity (%) |
|---|---|---|---|
| MCP | protein:vir:79157 protein:vir:79171 protein:vir:1153 | RSA1 phiE202 phi CTX | 67 % 66 % 61 % |
| Portal | protein:vir:79207 protein:vir:79150 protein:vir:1150 | phiE202 RSA1 phi CTX | 70 % 63 % 57 % |
| TermL | protein:vir:79177 protein:vir:1151 protein:vir:79128 | phiE202 phi CTX RSA1 | 73 % 65 % 64 % |
| MTP | protein:vir:79203 protein:vir:1173 protein:vir:79133 | phiE202 phi CTX RSA1 | 61 % 56 % 55 % |
| Sheath | protein:vir:79141 protein:vir:79181 protein:vir:1172 | RSA1 phiE202 phi CTX | 70 % 69 % 67 % |
| Ad1 | protein:vir:79188 protein:vir:79124 protein:vir:1155 | phiE202 RSA1 phi CTX | 53 % 44 % 42 % |
| Ne1 | protein:vir:79179 protein:vir:79115 protein:vir:1164 | phiE202 RSA1 phi CTX | 59 % 49 % 49 % |
| Tc1 | protein:vir:79169 protein:vir:1163 protein:vir:79131 | phiE202 phi CTX RSA1 | 49 % 47 % 45 % |