Welcome to InterEvData
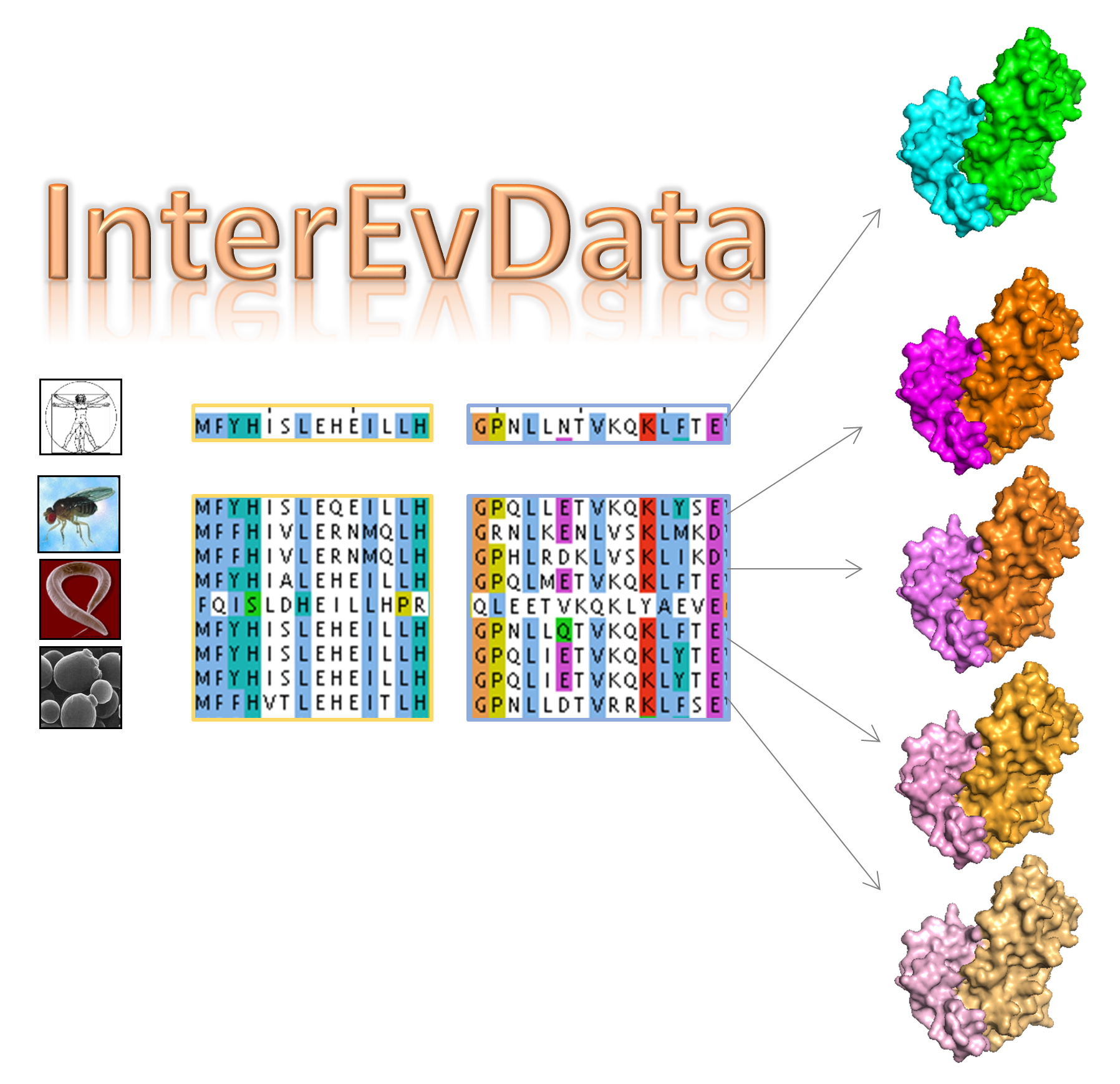
We share a novel way of combining scores with
evolutionary information at atomic level to improve near-native
discrimination rates in scoring. We make use of explicitely modelled
homologous decoys to enrich scoring of each query decoy by averaging the
scores over the query and its homologs. We illustrate this concept on
752 protein complexes from the PPI4DOCK benchmark (detailed list) and provide the
necessary data and a Singularity image to be able to reproduce the pipeline.
Download list:
We provide two datasets corresponding to input files and output results (scores and consensus) for the 752 PPI4DOCK cases used to benchmark our method, as well as 230 cases from the protein docking benchmark version 5 ("Weng" benchmark). For each dataset, we provide all unbound pdb files (query and modelled homology models), coMSAs for the two binding partners, the translation and rotation matrices form FRODOCK2.1, as well as output values for scores InterEvScore, SOAP-PP and Rosetta's Interface Score and their homology-enriched variants and consensuses.
-> PPI4DOCK_data.tar.gz (2.2 G, md5sum: 4b36da9747a8b5849860b3b594d45160)
-> Weng_BM5_data.tar.gz (805 M, md5sum: 421b9069e40ae2c3dc46ceb03907054c)
We also provide a Singularity image containing all tools and scripts necessary to execute the whole docking pipeline or generate or score decoy structures (InterEvData Code).
-> interevdata_tools.sif (770 M, md5sum: e67295e3bf384fd1845c6e392ab86046)
Short description:
The provided data aims at illustrating our novel concept of extrapolating evolutionary information contained in coMSAs to an atomic level of detail, making it directly compatible with multiple scoring functions in the light of molecular docking. Our example makes use of the 752 cases in the PPI4DOCK docking dataset for which all data and scripts necessary for application are provided.
Briefly, our method consists in modelling the unbound homologs identified in the coMSAs of both query protein partners based on the rapid threading protocol in RosettaCM. Once the query proteins are docked with FRODOCK2.1, homolog equivalents of each query decoy can be easily generated in a rigid-body fashion using FRODOCK's outputed rotation and translation docking coordinates, thus providing atomic models of homologous decoys directly associated with each query decoy without needing a computationally expensive clustering step. From then on, homolog decoys can be scored, just as query decoys, by a scoring function of your choice and we decided to average over the query and homolog scores of each decoy in order to get a final homology-based variant of our scoring function. In our example, we focus on three scores in addition to FRODOCK's mainly physics-based score: InterEvScore, which already implicitly takes co-evolutionary information into account in combination with a multi-body statistical potential; SOAP-PP, which uses a sophisticated atomic statistical potential; and Rosetta's Interface Score, which combines empirical and physical interaction terms.
How to cite us:
Chloé Quignot, Pierre Granger, Pablo Chacón, Raphaël Guerois and Jessica Andreani: Atomic-level evolutionary information improves protein-protein interface scoring (BioRxiv doi:10.1101/2020.10.26.355073)
Tools used:
FRODOCK2.1 (Ramírez-Aportela, E., et al. (2016). "FRODOCK 2.0: Fast Protein-Protein docking server." Bioinformatics: btw141. doi:10.1093/bioinformatics/btw141)
InterEvScore (Andreani, J., et al. (2013). "InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution." Bioinformatics 29(14): 1742-1749. doi:10.1093/bioinformatics/btt260)
SOAP-PP (Dong, G. Q., et al. (2013). "Optimized atomic statistical potentials: assessment of protein interfaces and loops." Bioinformatics 29(24): 3158-3166. doi: 10.1093/bioinformatics/btt560)
RosettaCM (Song, Y., Dimaio, F., Wang, R. Y., Kim, D., Miles, C., Brunette, T. J., … Baker, D. (2013). High-Resolution Comparative Modeling with RosettaCM. Structure, 21(SuppMat), 1735–42. doi:10.1016/j.str.2013.08.005)
Rosetta Interface Score (Lyskov, S. and J. J. Gray (2008). "The RosettaDock server for local protein-protein docking." Nucleic Acids Res 36(Web Server issue): W233-238. doi:10.1093/nar/gkn216; Chaudhury, S., et al. (2011). "Benchmarking and analysis of protein docking performance in Rosetta v3.2." PLoS One 6(8): e22477. doi:10.1371/journal.pone.0022477)
HHfilter (Steinegger, M., et al. (2019). "HH-suite3 for fast remote homology detection and deep protein annotation." BMC Bioinformatics 20(1): 473. doi:10.1186/s12859-019-3019-7)
Mafft (Katoh, K. and D. M. Standley (2013). "MAFFT multiple sequence alignment software version 7: improvements in performance and usability." Mol Biol Evol 30(4): 772-780. doi:10.1093/molbev/mst010)
Clustalw (Larkin, M. A., et al. (2007). "Clustal W and Clustal X version 2.0." Bioinformatics 23: 2947-2948. doi:10.1093/bioinformatics/btm404)
Singularity (Kurtzer, G.M., Sochat, V., Bauer, M.W. (2017). "Singularity: Scientific containers for mobility of compute." PLoS ONE 12(5): e0177459. doi:10.1371/journal.pone.0177459)
Download list:
We provide two datasets corresponding to input files and output results (scores and consensus) for the 752 PPI4DOCK cases used to benchmark our method, as well as 230 cases from the protein docking benchmark version 5 ("Weng" benchmark). For each dataset, we provide all unbound pdb files (query and modelled homology models), coMSAs for the two binding partners, the translation and rotation matrices form FRODOCK2.1, as well as output values for scores InterEvScore, SOAP-PP and Rosetta's Interface Score and their homology-enriched variants and consensuses.
-> PPI4DOCK_data.tar.gz (2.2 G, md5sum: 4b36da9747a8b5849860b3b594d45160)
- PPI4DOCK_list_752_at_least_one_FRODOCK_Acc_top10000_10plus_co_seqs_noAntibody.txt, a detailed list of the 752 PPI4DOCK cases used to benchmark our method (see FAQ question 2 for details on how to read this file)
- Five directories containing input data and results (see FAQ question 1 for content details)
- PPI4DOCK_pdbs/
- PPI4DOCK_MSA/
- PPI4DOCK_docking/
- PPI4DOCK_scores/
- PPI4DOCK_consensus/
-> Weng_BM5_data.tar.gz (805 M, md5sum: 421b9069e40ae2c3dc46ceb03907054c)
- info_BM5.txt, a list of the 230 protein docking benchmark v5 cases
- Five directories containing input data and results (see FAQ question 5 for content details)
- bm5_pdbs/
- bm5_MSA/
- bm5_docking/
- bm5_scores/
- bm5_consensus/
We also provide a Singularity image containing all tools and scripts necessary to execute the whole docking pipeline or generate or score decoy structures (InterEvData Code).
-> interevdata_tools.sif (770 M, md5sum: e67295e3bf384fd1845c6e392ab86046)
Short description:
The provided data aims at illustrating our novel concept of extrapolating evolutionary information contained in coMSAs to an atomic level of detail, making it directly compatible with multiple scoring functions in the light of molecular docking. Our example makes use of the 752 cases in the PPI4DOCK docking dataset for which all data and scripts necessary for application are provided.
Briefly, our method consists in modelling the unbound homologs identified in the coMSAs of both query protein partners based on the rapid threading protocol in RosettaCM. Once the query proteins are docked with FRODOCK2.1, homolog equivalents of each query decoy can be easily generated in a rigid-body fashion using FRODOCK's outputed rotation and translation docking coordinates, thus providing atomic models of homologous decoys directly associated with each query decoy without needing a computationally expensive clustering step. From then on, homolog decoys can be scored, just as query decoys, by a scoring function of your choice and we decided to average over the query and homolog scores of each decoy in order to get a final homology-based variant of our scoring function. In our example, we focus on three scores in addition to FRODOCK's mainly physics-based score: InterEvScore, which already implicitly takes co-evolutionary information into account in combination with a multi-body statistical potential; SOAP-PP, which uses a sophisticated atomic statistical potential; and Rosetta's Interface Score, which combines empirical and physical interaction terms.
How to cite us:
Chloé Quignot, Pierre Granger, Pablo Chacón, Raphaël Guerois and Jessica Andreani: Atomic-level evolutionary information improves protein-protein interface scoring (BioRxiv doi:10.1101/2020.10.26.355073)
Tools used:
FRODOCK2.1 (Ramírez-Aportela, E., et al. (2016). "FRODOCK 2.0: Fast Protein-Protein docking server." Bioinformatics: btw141. doi:10.1093/bioinformatics/btw141)
InterEvScore (Andreani, J., et al. (2013). "InterEvScore: a novel coarse-grained interface scoring function using a multi-body statistical potential coupled to evolution." Bioinformatics 29(14): 1742-1749. doi:10.1093/bioinformatics/btt260)
SOAP-PP (Dong, G. Q., et al. (2013). "Optimized atomic statistical potentials: assessment of protein interfaces and loops." Bioinformatics 29(24): 3158-3166. doi: 10.1093/bioinformatics/btt560)
RosettaCM (Song, Y., Dimaio, F., Wang, R. Y., Kim, D., Miles, C., Brunette, T. J., … Baker, D. (2013). High-Resolution Comparative Modeling with RosettaCM. Structure, 21(SuppMat), 1735–42. doi:10.1016/j.str.2013.08.005)
Rosetta Interface Score (Lyskov, S. and J. J. Gray (2008). "The RosettaDock server for local protein-protein docking." Nucleic Acids Res 36(Web Server issue): W233-238. doi:10.1093/nar/gkn216; Chaudhury, S., et al. (2011). "Benchmarking and analysis of protein docking performance in Rosetta v3.2." PLoS One 6(8): e22477. doi:10.1371/journal.pone.0022477)
HHfilter (Steinegger, M., et al. (2019). "HH-suite3 for fast remote homology detection and deep protein annotation." BMC Bioinformatics 20(1): 473. doi:10.1186/s12859-019-3019-7)
Mafft (Katoh, K. and D. M. Standley (2013). "MAFFT multiple sequence alignment software version 7: improvements in performance and usability." Mol Biol Evol 30(4): 772-780. doi:10.1093/molbev/mst010)
Clustalw (Larkin, M. A., et al. (2007). "Clustal W and Clustal X version 2.0." Bioinformatics 23: 2947-2948. doi:10.1093/bioinformatics/btm404)
Singularity (Kurtzer, G.M., Sochat, V., Bauer, M.W. (2017). "Singularity: Scientific containers for mobility of compute." PLoS ONE 12(5): e0177459. doi:10.1371/journal.pone.0177459)
