 |
ISPH: Iron-Sulfur ProteHome. |
 |
Dock3DinDepth: docking isosurfaces into AFM|SAXS|cryoEM envelopes. |
 |
CombineDocksAFM: combine of docking solutions of structural subunits under an AFM surface. |
 |
DockAFM: docking structures under an AFM surface. |
 |
ActinSimu: In silico study of actin-based force generation during cell movement or morphogenesis. |
 |
Morpheus offers a range of tools to analyze transcription factor binding sites (TFBS) on DNA sequences. |
 |
The InterEvol server is designed to combine structural and evolutionary analyses of protein complexes interfaces. |
 |
DeStripe uses a denoising protocol for the AFM images that are contaminated with heavy stripes. |
 |
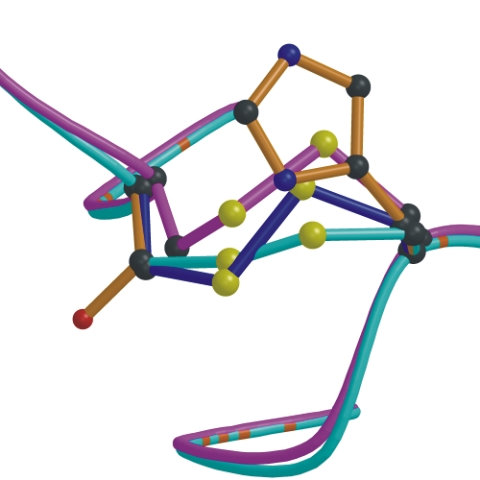
Adepth computes the atom depths in proteins which is derived from trilinear interpolation of the signed distance function (SDF) at the nearest grid points surrounding the atom. |
 |
BRIDGED server computes all the possibilities of inserting a disulfide bond into a protein. |
 |
The VIRFAM server can be used to analyze a set of sequences from bacteriophages genomes and check the existence of putative remote homologs of protein superfamilies (only recombinase so far) |
 |
P2CS is an integrated and comprehensive database of two-component system (TCS) signal transduction proteins, which contains a compilation of the TCS genes within 1125 completely sequenced genomes and 39 metagenomes. |
 |
P2TF is an integrated and comprehensive database of TF proteins, which contains a compilation of the TF genes within 1021 completely sequenced genomes and 39 metagenomes. |
 |
DSIR is a tool for siRNA (19 or 21 nt) and shRNA target design. |
 |
RASMOT-3D PRO searches in protein structure files for proteins possessing a group of residues in a topology similar to that adopted by a 3D motif given in input. |
 |
The STRIP server helps designing phospho-binding site mutants in S. cerevisiae proteins. |
 |
HMM-kalign package consists in the HMMER-2.3.2 package supplemented with a new function to generate sub-optimal alignments between one sequence and one HMM |
 |
The SCOTCH server can be used to score structural models of protein complexes based on evolutionary informations. |
 |
InteroPorc is an automatic prediction tool to infer protein-protein interaction networks. It is applicable for lots of species using orthology and known interactions. |



















