The neck is the head-to-tail interface which is formed by the connector and the tail-completion proteins. The connector is formed by the portal and the head-completion proteins. (Click here for a general description of the assembly pathway of tailed bacteriophages)
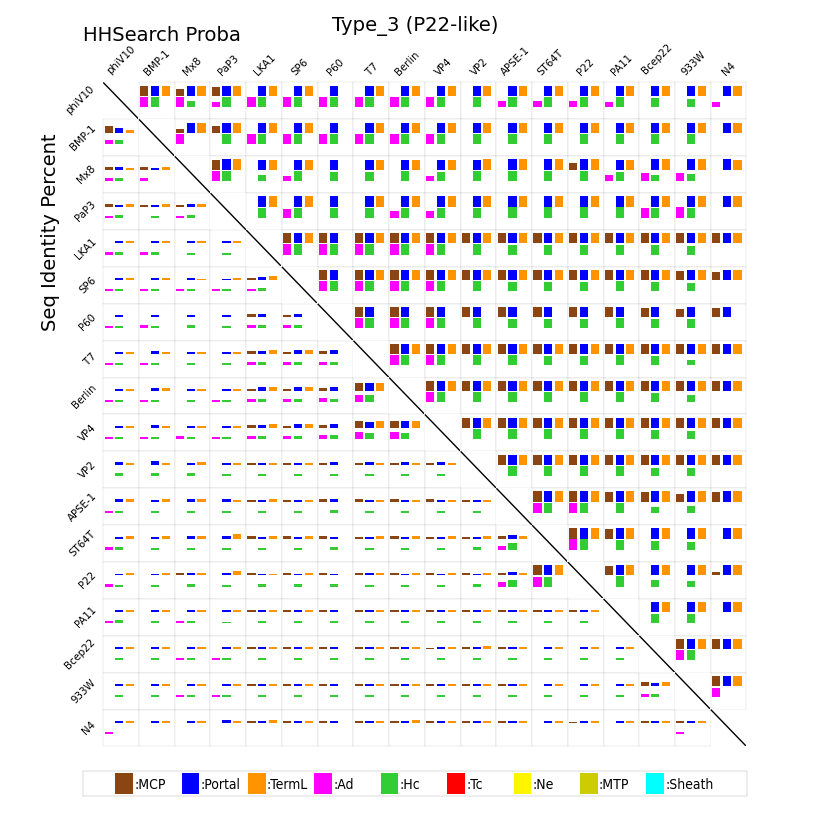
adopts the structural organization of the Podoviridae phage P22 neck. It is formed by a portal protein, an Ad3 (Adaptor of type 3), and a Hc3 (Head-completion of type 3) proteins with no tail-completion protein. Type 3 neck gathers podophages with genome sizes similar to those of siphophages (31 to 151 genes). In this neck organization, the phages can be divided into two groups according to their gene order. 31 phages over 52, exhibit a gene order that generally follows the [TermL-x(0-3)-Portal-x(1-4)-MCP-x(1-5)-Ad3-x(0-3)-Hc3] scheme. In two cases (119X and PaP2), Ad3 seems to be absent (or at least, has not been identified by our method). Most of the other phages (18 over 52) display a more conserved gene order (compared to the 31 previously mentioned phages) which generally follows the [Protal-x(1-4)-MCP-Ad3-Hc3-x(6-8)-TermL] scheme. The P60 phage seems to lack the TermL gene. The three remaining phages (N4, PBC5 and SIO1) are more atypical, lacking either both the Portal and MCP or the Hc3 genes and displaying more versatile gene order.


(-) indicates that the genetic context is given in the reverse order. For more clarity, numbers between square brackets indicate the number of unannotated genes.
For every pair of phages sharing a given neck type and for all the components of the capsid-neck-tail module they share (out of the nine possible components described below), we calculated the corresponding HHsearch probability and sequence identity scores. The resulting scores are illustrated with colored squares whose size is proportional to the considered score (similarity scores lie in the bottom half matrix while HHsearch probabilities lie in the top half matrix). See legend below for square colors and for identity or HHsearch probability scores.